Author
Lincoln Stein lstein@cshl.org lstein@cshl.org
Cold Spring Harbor Laboratory
Copyright
This document is copyright Lincoln Stein, 2002. It can be copied and distributed under the terms of the Perl Artistic License.
Revision History
| Revision | About |
|---|---|
| 0.1 2002-09-01 | First release |
| 0.2 2003-05-15 lds | Current as of BioPerl 1.2.2 |
| 0.3 Torsten Seemann 06:17, 22 December 2005 (EST) | BioPerl Wiki & fixed figure references |
| 0.4 Jason Stajich 13:11, 28 December 2005 (EST) | Added many links to modules |
| 0.5 Mauricio Herrera 14:56, 29 October 2006 (EST) | Added syntax highlighting and code formatting |
Abstract
This HOWTO describes how to render sequence data graphically in a horizontal map. It applies to a variety of situations ranging from rendering the feature table of a GenBank entry, to graphing the positions and scores of a BLAST search, to rendering a clone map. It describes the programmatic interface to the module, and discusses how to create dynamic web pages using and the GBrowse package.
Introduction
This HOWTO describes the Bio::Graphics module, and some of the applications that were built on top of it. Bio::Graphics was designed to solve the following common problems:
- You have a list of BLAST hits on a sequence and you want to generate a picture that shows where the hits go and what their score is.
- You have a big GenBank file with a complex feature table, and you want to render the positions of the genes, repeats, promoters and other features.
- You have a list of ESTs that you’ve mapped to a genome, and you want to show how they align.
- You have created a clone fingerprint map, and you want to display it.
The Bio::Graphics module was designed to solve these problems. In addition, using the Bio::Graphics module (part of BioPerl) and the GBrowse program (available from http://www.gmod.org) you can create interactive web pages to explore your data.
This document takes you through a few common applications of Bio::Graphics in a cookbook fashion. Advanced users might be interested in the Custom Glyph HOWTO, which explains how to extend Bio::Graphics using custom glyphs.
Preliminaries
Bio::Graphics is dependent on GD, a Perl module for generating bitmapped graphics written by the author. GD in turn is dependent on libgd, a C library written by Thomas Boutell, formerly also of Cold Spring Harbor Laboratory. To use Bio::Graphics you must have both these software libraries installed.
If you are on a Linux system, you might already have GD installed. To verify, run the following command:
% perl -MGD -e 'print $GD::VERSION, "\n"';
If the program prints out a version number, you are in luck. Otherwise, if you get a Can't locate GD.pm error, you’ll have to install the module. For users of ActiveState Perl this is very easy. Just start up the PPM program and issue the command install GD. For users of other versions of Perl, you should go to www.cpan.org, download a recent version of the module, unpack it, and follow the installation instructions.
You may need to upgrade to a recent version of the libgd C library.
Getting Started
All the code examples, BLAST input files, and sequence files we’ll use are available here:
- BLASTN output
- render_blast1.pl
- render_blast2.pl
- render_blast3.pl
- render_blast4.pl
- embl2picture.pl
- render_features.pl
- factor7.embl
Our first example will be rendering a table of BLAST hits on a sequence that is exactly 1000 residues long. For now, we’re ignoring finicky little details like HSPs, and assume that each hit is a single span from start to end. Also, we’ll be using the BLAST score rather than P or E value. Later on, we’ll switch to using real BLAST output parsed by the module, but for now, our table looks like this:
Table 1. Simple blast hit file (data1.txt)
# hit score start end
hsHOX3 381 2 200
scHOX3 210 2 210
xlHOX3 800 2 200
hsHOX2 1000 380 921
scHOX2 812 402 972
xlHOX2 1200 400 970
BUM 400 300 620
PRES1 127 310 700
Hint when copy-pasting this into your own text, make sure that there are tab stops between the different columns. You can use your favorite text editor to replace the spaces to tabs.
Our first attempt to parse and render this file looks like this:
Example 1. Rendering the simple blast hit file (render_blast1.pl)
#!/usr/bin/perl
# This is code example 1 in the Graphics-HOWTO
use strict; use Bio::Graphics; use Bio::SeqFeature::Generic;
my $panel = Bio::Graphics::Panel->new(
-length => 1000,
-width => 800
);
my $track = $panel->add_track(
-glyph => 'generic',
-label => 1
);
while (<>) { # read blast file
chomp;
next if /^#/; # ignore comments
my($name,$score,$start,$end) = split /\t+/;
my $feature = Bio::SeqFeature::Generic->new(
-display_name => $name,
-score => $score,
-start => $start,
-end => $end
);
$track->add_feature($feature);
}
print $panel->png;
The script begins by loading the module (line 3), which in turn brings in a number of other modules that we’ll use later. We also load in order to create a series of objects for rendering. We then create a object by calling its new() method, specifying that the panel is to correspond to a sequence that is 1000 nucleotides long, and has a physical width of 800 pixels (line 5). The Panel can contain multiple horizontal tracks, each of which has its own way of rendering features (called a “glyph”), color, labeling convention, and so forth. In this simple example, we create a single track by calling the panel object’s add_track() method (line 6), specify a glyph type of “generic”, and ask that the objects in the track be labeled by providing a true value to the -label argument. This gives us a track object that we can add our hits to.
We’re now ready to render the BLAST hit file. We loop through it (line 7-14), stripping off the comments, and parsing out the name, score and range (line 10). We now need a object to place in the track. The easiest way to do this is to create a Bio::SeqFeature::Generic object, which is similar to Bio::PrimarySeq, except that it provides a way of attaching start and end positions to the sequence, as well as such nebulous but useful attributes as the “score” and “source”. The Bio::SeqFeature::Generic->new() method, invoked in line 11, takes arguments corresponding to the name of each hit, its start and end coordinates, and its score.
After creating the feature object, we add it to the track by calling the track’s add_feature() method (line 13).
After processing all the hits, we call the panel’s png() method to render them and convert it into a Portable Network Graphics file, the contents of which are printed to standard output. We can now view the result by piping it to our favorite image display program. Users of operating systems that don’t support pipes can simply redirect the output to a file and view it in their favorite image program.
% perl render_blast1.pl data1.txt | display -
Figure 1. Rendering BLAST hits 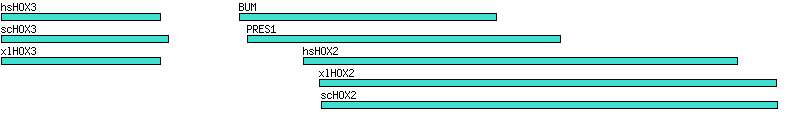
Important!
If you are on a Windows platform, you need to put STDOUT into binary
mode so that the PNG file does not go through Window's carriage
return/linefeed transformations. Before the final print statement, put
the statement binmode(STDOUT).
Adding a Scale to the Image
This is all very nice, but it’s missing two essential components:
- It doesn’t have a scale.
- It doesn’t distinguish between hits with different scores.
Example 2 fixes these problems.
Example 2. Rendering the blast hit file with scores and scale
#!/usr/bin/perl
# This is code example 2 in the Graphics-HOWTO
use strict;
use lib '/home/lstein/projects/bioperl-live';
use Bio::Graphics;
use Bio::SeqFeature::Generic;
my $panel = Bio::Graphics::Panel->new(
-length => 1000,
-width => 800,
-pad_left => 10,
-pad_right => 10,
);
my $full_length = Bio::SeqFeature::Generic->new(
-start => 1,
-end => 1000
);
$panel->add_track($full_length,
-glyph => 'arrow',
-tick => 2,
-fgcolor => 'black',
-double => 1
);
my $track = $panel->add_track(
-glyph => 'graded_segments',
-label => 1,
-bgcolor => 'blue',
-min_score => 0,
-max_score => 1000
);
while (<>) { # read blast file
chomp;
next if /^#/; # ignore comments
my($name,$score,$start,$end) = split /\t+/;
my $feature = Bio::SeqFeature::Generic->new(
-display_name => $name,
-score => $score,
-start => $start,
-end => $end
);
$track->add_feature($feature);
}
print $panel->png;
There are several changes to look at. The first is minor. We’d like to put a boundary around the left and right edges of the image so that the features don’t bump up against the margin, so we specify a 10 pixel leeway with the -pad_left and -pad_right arguments in lines 8 and 9.
The next change is more subtle. We want to draw a scale all the way across the image. To do this, we create a track to contain the scale, and a feature that spans the track from the start to the end. Line 11 creates the feature, giving its start and end coordinates. Lines 12-17 create a new track containing this feature. Unlike the previous example, in which we created the track first and then added features one at a time with add_feature(), we show here how to add feature(s) directly in the call to add_track(). If the first argument to add_track is either a single feature or a feature array ref, then add_track() will automatically incorporate the feature(s) into the track in a single efficient step. The remainder of the arguments configure the track as before. The -glyph argument says to use the “arrow” glyph. The -tick argument indicates that the arrow should contain tick marks, and that both major and minor ticks should be shown (tick type of “2”). We set the foreground color to black, and request that arrows should be placed at both ends (-double =>1).
In lines 18-22, we get a bit fancier with the blast hit track. Now, instead of creating a generic glyph, we use the “graded_segments” glyph. This glyph takes the specified background color for the feature, and either darkens or lightens it according to its score. We specify the base background color (-bgcolor => 'blue'), and the minimum and maximum scores to scale to (-min_score and -max_score). (You may need to experiment with the min and max scores in order to get the glyph to scale the colors the way you want.) The remainder of the program is the same.
When we run the modified script, we get this image:
Figure 2. The improved image. 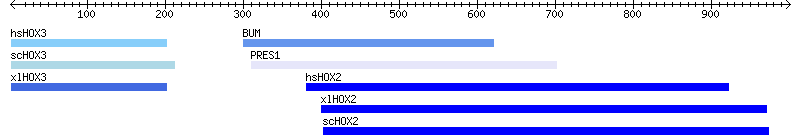
Important!
` Remember that if you are on a Windows platform, you need to put STDOUT
into binary mode so that the PNG file does not go through Window’s
carriage return/linefeed transformations. Before the final print
statement, write binmode(STDOUT).`
Improving the Image
Before we move into displaying gapped alignments, let’s tweak the image slightly so that higher scoring hits appear at the top of the image, and the score itself is printed in red underneath each hit. The changes are shown in Example 3.
Example 3. Rendering the blast hit file with scores and scale
#!/usr/bin/perl
# This is code example 3 in the Graphics-HOWTO
use strict;
use lib '/home/lstein/projects/bioperl-live';
use Bio::Graphics; use Bio::SeqFeature::Generic;
my $panel = Bio::Graphics::Panel->new(
-length => 1000,
-width => 800,
-pad_left => 10,
-pad_right => 10,
);
my $full_length = Bio::SeqFeature::Generic->new(
-start => 1,
-end => 1000,
);
$panel->add_track($full_length,
-glyph => 'arrow',
-tick => 2,
-fgcolor => 'black',
-double => 1,
);
my $track = $panel->add_track(
-glyph => 'graded_segments',
-label => 1,
-bgcolor => 'blue',
-min_score => 0,
-max_score => 1000,
-font2color => 'red',
-sort_order => 'high_score',
-description => sub {
my $feature = shift;
my $score = $feature->score;
return "score=$score";
},
);
while (<>) { # read blast file
chomp;
next if /^#/; # ignore comments
my($name,$score,$start,$end) = split /\t+/;
my $feature = Bio::SeqFeature::Generic->new(
-score => $score,
-display_name => $name,
-start => $start,
-end => $end,
);
$track->add_feature($feature);
}
print $panel->png;
There are two changes to look at. The first appears in line 24, where we pass the -sort_order option to the call that creates the blast hit track. -sort_order changes the way that features sort from top to bottom, and will accept a number of prepackaged sort orders or a coderef for custom sorting. In this case, we pass a prepackaged sort order of high_score, which sorts the hits from top to bottom in reverse order of their score.
The second change is more complicated, and involves the -description option that appears in the add_track() call on lines 25-28. The value of -description will be printed beneath each feature. We could pass -description a constant string, but that would simply print the same string under each feature. Instead we pass -description a code reference to a subroutine that will be invoked while the picture is being rendered. This subroutine will be passed the current feature, and must return the string to use as the value of the description. In our code, we simply fetch out the BLAST hit’s score using its score() method, and incorporate that into the description string.
Tip:
` The ability to use a code reference as a configuration option isn’t
unique to -description. In fact, you can use a code reference for any
of the options passed to add_track().`
Another minor change is the use of -font2color in line 23. This simply sets the color used for the description strings. Figure 3 shows the effect of these changes.
Figure 3. The image with descriptions and sorted hits 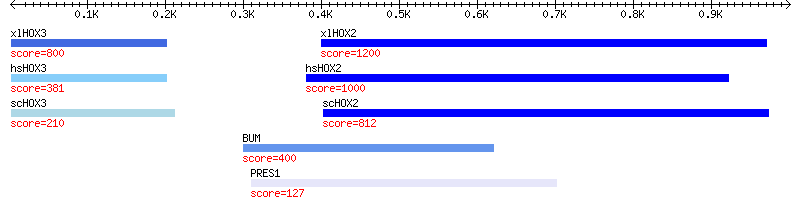
Parsing Real BLAST Output
From here it’s just a small step to writing a general purpose utility that will read a BLAST file, parse its output, and output a picture. The key is to use the infrastructure because it produces similarity hits that can be rendered directly. Code example 4 shows the new utility.
Example 4. Parsing and Rendering a Real BLAST File with Bio::SearchIO
#!/usr/bin/perl
# This is code example 4 in the Graphics-HOWTO
use strict;
use lib "$ENV{HOME}/projects/bioperl-live";
use Bio::Graphics;
use Bio::SearchIO;
use Bio::SeqFeature::Generic;
my $file = shift or die "Usage: render_blast4.pl <blast file>\n";
my $searchio = Bio::SearchIO->new(-file => $file,
-format => 'blast') or die "parse failed";
my $result = $searchio->next_result() or die "no result";
my $panel = Bio::Graphics::Panel->new(
-length => $result->query_length,
-width => 800,
-pad_left => 10,
-pad_right => 10,
);
my $full_length = Bio::SeqFeature::Generic->new(
-start => 1,
-end => $result->query_length,
-display_name => $result->query_name,
);
$panel->add_track($full_length,
-glyph => 'arrow',
-tick => 2,
-fgcolor => 'black',
-double => 1,
-label => 1,
);
my $track = $panel->add_track(
-glyph => 'graded_segments',
-label => 1,
-connector => 'dashed',
-bgcolor => 'blue',
-font2color => 'red',
-sort_order => 'high_score',
-description => sub {
my $feature = shift;
return unless $feature->has_tag('description');
my ($description) = $feature->each_tag_value('description');
my $score = $feature->score;
"$description, score=$score";
},
);
while( my $hit = $result->next_hit ) {
next unless $hit->significance < 1E-20;
my $feature = Bio::SeqFeature::Generic->new(
-score => $hit->raw_score,
-display_name => $hit->name,
-tag => {
description => $hit->description
},
);
while ( my $hsp = $hit->next_hsp ) {
$feature->add_sub_SeqFeature($hsp,'EXPAND');
}
$track->add_feature($feature);
}
print $panel->png;
In lines 6-8 we read the name of the file that contains the BLAST results from the command line, and pass it to Bio::SearchIO->new(), returning a parser object. We read a single result from the searchIO object (line 9). This assumes that the BLAST output file contains a single run of BLAST only.
We then initialize the panel and tracks as before. The only change here is in lines 24-36, where we create the track for the BLAST hits. The -description option has now been enhanced to create a line of text that incorporates the “description” tag from the feature object as well as its similarity score. There’s also a slight change in line 26, where we introduce the -connector option. This allows us to configure a line that connects the segments of a discontinuous feature, such as the HSPs in a BLAST hit. In this case, we asked the rendering engine to produce a dashed connector line.
The remainder of the script retrieves each of the hits from the BLAST file, creates a Feature object representing the hit, and then retrieves each HSP and incorporates it into the feature. Line 37 begins a while() loop that retrieves each of the similarity hits in turn. We filter the hit by its significance, throwing out any that have an expectation value greater than 1E-20 (you will have to adjust this in your own utilities). We then use the information in the hit to construct a object (lines 39-44). Notice how the name of the hit and the score are used to initialize the feature, and how the description is turned into a tag named “description.”
The start and end bounds of the hit are determined by the union of its HSPs. We loop through each of the hit’s HSPs by calling its next_hsp() method, and add each HSP to the newly-created hit feature by calling the feature’s add_sub_SeqFeature() method (line 46). The EXPAND parameter instructs the feature to expand its start and end coordinates to enclose the added subfeature.
Once all the HSPs are added to the feature, we insert the feature into the track as before using the track’s add_feature() function.
Figure 4 shows the output from a sample BLAST hit file.
Figure 4. Output from the BLAST parsing and rendering script 
The next section will demonstrate how to parse and display feature tables from GenBank and EMBL.
Important!
Remember that if you are on a Windows platform, you need to put STDOUT
` into binary mode so that the PNG file does not go through Window’s
carriage return/linefeed transformations. Before the final print
statement, write binmode(STDOUT).`
Rendering Features from a GenBank or EMBL File
With you can render the feature table of a GenBank or EMBL file quite easily. The trick is to use to generate a set of objects, and to use those features to populate tracks (see the Feature-Annotation HOWTO for more information on features). The documentation for each of the individual. For simplicity’s sake, we will sort each feature by its primary tag (such as “exon”) and create a new track for each tag type. Code example 5 shows the code for rendering an EMBL or GenBank entry.
Example 5. The embl2picture.pl script turns any EMBL or GenBank entry into a graphical rendering.
#!/usr/bin/perl
# file: embl2picture.pl
# This is code example 5 in the Graphics-HOWTO
# Author: Lincoln Stein
use strict;
use lib "$ENV{HOME}/projects/bioperl-live";
use Bio::Graphics;
use Bio::SeqIO;
use Bio::SeqFeature::Generic;
my $file = shift or die "provide a sequence file as the argument";
my $io = Bio::SeqIO->new(-file=>$file) or die "couldn't create Bio::SeqIO";
my $seq = $io->next_seq or die "couldn't find a sequence in the file";
my $wholeseq = Bio::SeqFeature::Generic->new(
-start => 1,
-end => $seq->length,
-display_name => $seq->display_name
);
my @features = $seq->all_SeqFeatures;
# partition features by their primary tags
my %sorted_features; for my $f (@features) {
my $tag = $f->primary_tag;
push @{$sorted_features{$tag}},$f;
}
my $panel = Bio::Graphics::Panel->new(
-length => $seq->length,
-key_style => 'between',
-width => 800,
-pad_left => 10,
-pad_right => 10,
);
$panel->add_track($wholeseq,
-glyph => 'arrow',
-bump => 0,
-double => 1,
-tick => 2);
$panel->add_track($wholeseq,
-glyph => 'generic',
-bgcolor => 'blue',
-label => 1,
);
# general case
my @colors = qw(cyan orange blue purple green
chartreuse magenta yellow aqua);
my $idx = 0;
for my $tag (sort keys %sorted_features) {
my $features = $sorted_features{$tag};
$panel->add_track($features,
-glyph => 'generic',
-bgcolor => $colors[$idx++ % @colors],
-fgcolor => 'black',
-font2color => 'red',
-key => "${tag}s",
-bump => +1,
-height => 8,
-label => 1,
-description => 1,
);
}
print $panel->png;
exit 0;
The way this script works is simple. After the library load preamble, the script reads the name of the GenBank or [EMBL] file from the command line (line 8). It passes the filename to ‘s new() method, and reads the first sequence object from it (lines 9-11). If anything goes wrong, the script dies with an error message.
The returned object is a object, which has a length but no defined start or end coordinates. We would like to create a drawable object to use for the scale, so we generate a new object that goes from a start of 1 to the length of the sequence. (lines 12-13).
The script reads the features from the sequence object by calling all_SeqFeatures(), and then sorts each feature by its primary tag into a hash of array references named %sorted_features (lines 14-20).
Next, we create the object (lines 21-27). As in previous examples, we specify the width of the image, as well as some extra white space to pad out the left and right borders.
We now add two tracks, one for the scale (lines 28-32) and the other for the sequence as a whole (33-37). As in the earlier examples, we pass add_track() the sequence object as the first argument before the options so that the object is incorporated into the track immediately.
We are now ready to create a track for each feature type. In order to distinguish the tracks by color, we initialize an array of 9 color names and simply cycle through them (lines 39-54). For each feature tag, we retrieve the corresponding list of features from %sorted_features (line 42) and create a track for it using the “generic” glyph and the next color in the list (lines 43-53). We set the -label and -description options to the value “1”. This signals that it should do the best it can to choose useful label and description values on its own.
After adding all the feature types, we call the panel’s png() method to generate a graphic file, which we print to STDOUT. If we are on a Windows platform, we would have to include binmode(STDOUT) prior to this statement in order to avoid Windows textmode carriage return/linefeed transformations. Figure 5 shows an example of the output of this script using an EMBL sequence file as input.
Figure 5. Example output image. 
A Better Version of the Feature Renderer
The previous example’s rendering has numerous deficiencies. For one thing, there are no lines connecting the various CDS rectangles in the CDS track to show how they are organized into a spliced transcript. For another, the repetition of the source tag “EMBL/GenBank/SwissProt” is not particularly illuminating.
However, it’s quite easy to customize the display, making the script into a generally useful utility. The revised code is shown in example 6.
Example 6. The embl2picture.pl script turns any EMBL or GenBank entry into a graphical rendering
#!/usr/bin/perl
# file: embl2picture.pl
# This is code example 6 in the Graphics-HOWTO
# Author: Lincoln Stein
use strict;
use lib "$ENV{HOME}/projects/bioperl-live";
use Bio::Graphics;
use Bio::SeqIO;
use Bio::SeqFeature::Generic;
use constant USAGE =><<END;
Usage: $0 <file>
Render a GenBank/EMBL entry into drawable form.
Return as a GIF or PNG image on standard output.
File must be in embl, genbank, or another SeqIO-
recognized format. Only the first entry will be
rendered.
Example to try:
embl2picture.pl factor7.embl | display -
END
my $file = shift or die USAGE;
my $io = Bio::SeqIO->new(-file=>$file) or die USAGE;
my $seq = $io->next_seq or die USAGE;
my $wholeseq = Bio::SeqFeature::Generic->new(
-start => 1,
-end => $seq->length,
-display_name => $seq->display_name
);
my @features = $seq->all_SeqFeatures;
# sort features by their primary tags
my %sorted_features;
for my $f (@features) {
my $tag = $f->primary_tag;
push @{$sorted_features{$tag}},$f;
}
my $panel = Bio::Graphics::Panel->new(
-length => $seq->length,
-key_style => 'between',
-width => 800,
-pad_left => 10,
-pad_right => 10,
);
$panel->add_track($wholeseq,
-glyph => 'arrow',
-bump => 0,
-double => 1,
-tick => 2,
);
$panel->add_track($wholeseq,
-glyph => 'generic',
-bgcolor => 'blue',
-label => 1,
);
$gene_label = gene_label();
$gene_label = gene_label();
$generic_description = generic_description();
# special cases
if ($sorted_features{CDS}) {
$panel->add_track($sorted_features{CDS},
-glyph => 'transcript2',
-bgcolor => 'orange',
-fgcolor => 'black',
-font2color => 'red',
-key => 'CDS',
-bump => +1,
-height => 12,
-label => $gene_label,
-description => $gene_description,
);
delete $sorted_features{'CDS'};
}
if ($sorted_features{tRNA}) {
$panel->add_track($sorted_features{tRNA},
-glyph => 'transcript2',
-bgcolor => 'red',
-fgcolor => 'black',
-font2color => 'red',
-key => 'tRNAs',
-bump => +1,
-height => 12,
-label => $gene_label,
);
delete $sorted_features{tRNA};
}
# general case
my @colors = qw(cyan orange blue purple green
chartreuse magenta yellow aqua);
my $idx = 0;
for my $tag (sort keys %sorted_features) {
my $features = $sorted_features{$tag};
$panel->add_track($features,
-glyph => 'generic',
-bgcolor => $colors[$idx++ % @colors],
-fgcolor => 'black',
-font2color => 'red',
-key => "${tag}s",
-bump => +1,
-height => 8,
-description => $generic_description,
);
}
print $panel->png; exit 0;
sub gene_label {
my $feature = shift;
my @notes;
foreach (qw(product gene)) {
@notes = eval {$feature->get_tag_values($_)};
last;
}
$notes[0];
}
sub gene_description {
my $feature = shift;
my @notes;
foreach (qw(note)) {
@notes = eval{$feature->get_tag_values($_)};
last;
}
return unless @notes;
substr($notes[0],30) = '...' if length $notes[0] > 30;
$notes[0];
}
sub generic_description {
my $feature = shift;
my $description;
foreach ($feature->get_all_tags) {
my @values = $feature->get_tag_values($_);
$description .= $_ eq 'note' ? "@values" : "$_=@values; ";
}
$description =~ s/; $//; # get rid of last
$description;
}
At 124 lines, this is the longest example in this HOWTO, but the changes are straightforward. The major difference occurs in lines 47-61 and 62-74, where we handle two special cases: “CDS” records and “tRNAs”. For these two feature types we would like to draw the features like genes using the “transcript2” glyph. This glyph draws inverted V’s for introns, if there are any, and will turn the last (or only) exon into an arrow to indicate the direction of transcription.
First we look to see whether there are any features with the primary tag of “CDS” (lines 47-61). If so, we create a track for them using the desired glyph. Line 49 shows how to add several features to a track at creation time. If the first argument to add_track() is an array reference, all the features contained in the array will be incorporated into the track. We provide custom code references for the -label and -description options. As we shall see later, the subroutines these code references point to are responsible for extracting names and descriptions for the coding regions. After we handle this special case, we remove the CDS feature type from the %sorted_features associative array.
We do the same thing for tRNA features, but with a different color scheme (lines 62-74).
Having dealt with the special cases, we render the remaining feature types using the same code we used earlier. The only change is that instead of allowing to guess at the description from the feature’s source tag, we use the -description option to point to a subroutine that will generate more informative description strings.
The gene_label() (lines 93-102) and gene_description() (lines 103-114) subroutines are simple. The first one searches the feature for the tags “product” and/or “gene” and uses the first one it finds as the label for the feature. The gene_description() subroutine is similar, except that it returns the value of the first tag named “note”. If the description is over 30 characters long, it is truncated. Notice that we place calls to get_tag_values() inside eval{} blocks in order to avoid having an exception raised if the feature does not have a tag with the desired value.
The generic_description() (lines 115-124) is invoked to generate descriptions of all non-gene features. We simply concatenate together the names and values of tags. For example the entry:
source 1..12850
/db_xref="taxon:9606"
/organism="Homo sapiens"
/map="13q34"
will be turned into the description string "db_xref=taxon:9606; organism=Homo Sapiens; map=13q34
After adding all the feature types, we call the panel’s png() method to generate a graphic file, which we print to STDOUT. Figure 6 shows an example of the output of this script.
Figure 6. Example output with connected CDS. 
Summary
In summary, we have seen how to use the module to generate representations of sequence features as horizontal maps. We applied these techniques to two common problems: rendering the output of a BLAST run, and rendering the feature table of a GenBank/EMBL entry.
The graphics module is quite flexible. In addition to the options that we have seen, there are glyphs for generating point-like features such as SNPs, specialized glyphs that draw GC content and open reading frames, and glyphs that generate histograms, bar charts and other types of graphs. has been used to represent physical (clone) maps, radiation hybrid maps, EST clusters, cytogenetic maps, restriction maps, and much more.
Although we haven’t shown it, provides support for generating HTML image maps. The Generic Genome Browser uses this facility to generate clickable, browsable images of the genome from a variety of genome databases.
Another application you should investigate is the script. This script uses the BioFetch interface to fetch GenBank/EMBL/SwissProt entries dynamically from the web before rendering them into PNG images.
Finally, if you find yourself constantly tweaking the graphic options, you might be interested in , a utility module for interpreting and rendering a simple tab-delimited format for sequence features. is a Perl script built on top of this module, which you can find in the scripts/graphics directory in the BioPerl distribution.
Tip Obtain the list of glyphs by running perldoc on . You can also obtain a description of the glyph options by running perldoc on individual glyphs, for example, for Bio::Graphics::Glyph::arrow:
% perldoc Bio::Graphics::Glyph::arrow